Example of EGMS toolkit in Python environment
Date: 26 Jan. 2024 by Alexis Hrysiewicz
Import the required modules
As a Python package, we import the EGMS-toolkit sub-packages. For this example, (and for a complete processing), EGMS-toolkit requires 4 modules:
EGMSS1burstIDapi: management of the S1-Burst-ID map (from the classes modules);
EGMSS1ROIapi: process the selection of datasets based on a ROI (from the classes modules);
EGMSdownloaderapi: downloader of EGMS data (from the classes modules);
egmsdatatools: some supplementary tools (from the functions modules).
[1]:
from EGMStoolkit.classes import EGMSS1burstIDapi
from EGMStoolkit.classes import EGMSS1ROIapi
from EGMStoolkit.classes import EGMSdownloaderapi
from EGMStoolkit.functions import egmsdatatools
Of course, it also is possible to directly import EGMStoolkit:
import EGMStoolkit
Manage the Sentinel-1-Burst-ID map
The first step is the creation of the S1burstIDmap object to manage the Sentinel-1-Burst-ID map.
NOTE: To improve the clarity of this example, the verbose mode will be actived for some selected processing step.
Create the python variable
We call the class with the verbose and logging modes deactived.
[2]:
info = EGMSS1burstIDapi.S1burstIDmap(verbose=False,log=None)
Of each EGMS-toolkit class, the print() method can be used to visualise its attributes.
[3]:
info.print()
date_str_init: 29/05/2022, dirmap: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/3rdparty, pathIDmap: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/3rdparty/S1_burstid_20220530, list_date: ['20220530', '20220531', '20220601', '20220602', '20220603', '20220604', '20220605', '20220606', '20220607', '20220608', '20220609', '20220610', '20220611', '20220612', '20220613', '20220614', '20220615', '20220616', '20220617', '20220618', '20220619', '20220620', '20220621', '20220622', '20220623', '20220624', '20220625', '20220626', '20220627', '20220628', '20220629', '20220630', '20220701', '20220702', '20220703', '20220704', '20220705', '20220706', '20220707', '20220708', '20220709', '20220710', '20220711', '20220712', '20220713', '20220714', '20220715', '20220716', '20220717', '20220718', '20220719', '20220720', '20220721', '20220722', '20220723', '20220724', '20220725', '20220726', '20220727', '20220728', '20220729', '20220730', '20220731', '20220801', '20220802', '20220803', '20220804', '20220805', '20220806', '20220807', '20220808', '20220809', '20220810', '20220811', '20220812', '20220813', '20220814', '20220815', '20220816', '20220817', '20220818', '20220819', '20220820', '20220821', '20220822', '20220823', '20220824', '20220825', '20220826', '20220827', '20220828', '20220829', '20220830', '20220831', '20220901', '20220902', '20220903', '20220904', '20220905', '20220906', '20220907', '20220908', '20220909', '20220910', '20220911', '20220912', '20220913', '20220914', '20220915', '20220916', '20220917', '20220918', '20220919', '20220920', '20220921', '20220922', '20220923', '20220924', '20220925', '20220926', '20220927', '20220928', '20220929', '20220930', '20221001', '20221002', '20221003', '20221004', '20221005', '20221006', '20221007', '20221008', '20221009', '20221010', '20221011', '20221012', '20221013', '20221014', '20221015', '20221016', '20221017', '20221018', '20221019', '20221020', '20221021', '20221022', '20221023', '20221024', '20221025', '20221026', '20221027', '20221028', '20221029', '20221030', '20221031', '20221101', '20221102', '20221103', '20221104', '20221105', '20221106', '20221107', '20221108', '20221109', '20221110', '20221111', '20221112', '20221113', '20221114', '20221115', '20221116', '20221117', '20221118', '20221119', '20221120', '20221121', '20221122', '20221123', '20221124', '20221125', '20221126', '20221127', '20221128', '20221129', '20221130', '20221201', '20221202', '20221203', '20221204', '20221205', '20221206', '20221207', '20221208', '20221209', '20221210', '20221211', '20221212', '20221213', '20221214', '20221215', '20221216', '20221217', '20221218', '20221219', '20221220', '20221221', '20221222', '20221223', '20221224', '20221225', '20221226', '20221227', '20221228', '20221229', '20221230', '20221231', '20230101', '20230102', '20230103', '20230104', '20230105', '20230106', '20230107', '20230108', '20230109', '20230110', '20230111', '20230112', '20230113', '20230114', '20230115', '20230116', '20230117', '20230118', '20230119', '20230120', '20230121', '20230122', '20230123', '20230124', '20230125', '20230126', '20230127', '20230128', '20230129', '20230130', '20230131', '20230201', '20230202', '20230203', '20230204', '20230205', '20230206', '20230207', '20230208', '20230209', '20230210', '20230211', '20230212', '20230213', '20230214', '20230215', '20230216', '20230217', '20230218', '20230219', '20230220', '20230221', '20230222', '20230223', '20230224', '20230225', '20230226', '20230227', '20230228', '20230301', '20230302', '20230303', '20230304', '20230305', '20230306', '20230307', '20230308', '20230309', '20230310', '20230311', '20230312', '20230313', '20230314', '20230315', '20230316', '20230317', '20230318', '20230319', '20230320', '20230321', '20230322', '20230323', '20230324', '20230325', '20230326', '20230327', '20230328', '20230329', '20230330', '20230331', '20230401', '20230402', '20230403', '20230404', '20230405', '20230406', '20230407', '20230408', '20230409', '20230410', '20230411', '20230412', '20230413', '20230414', '20230415', '20230416', '20230417', '20230418', '20230419', '20230420', '20230421', '20230422', '20230423', '20230424', '20230425', '20230426', '20230427', '20230428', '20230429', '20230430', '20230501', '20230502', '20230503', '20230504', '20230505', '20230506', '20230507', '20230508', '20230509', '20230510', '20230511', '20230512', '20230513', '20230514', '20230515', '20230516', '20230517', '20230518', '20230519', '20230520', '20230521', '20230522', '20230523', '20230524', '20230525', '20230526', '20230527', '20230528', '20230529', '20230530', '20230531', '20230601', '20230602', '20230603', '20230604', '20230605', '20230606', '20230607', '20230608', '20230609', '20230610', '20230611', '20230612', '20230613', '20230614', '20230615', '20230616', '20230617', '20230618', '20230619', '20230620', '20230621', '20230622', '20230623', '20230624', '20230625', '20230626', '20230627', '20230628', '20230629', '20230630', '20230701', '20230702', '20230703', '20230704', '20230705', '20230706', '20230707', '20230708', '20230709', '20230710', '20230711', '20230712', '20230713', '20230714', '20230715', '20230716', '20230717', '20230718', '20230719', '20230720', '20230721', '20230722', '20230723', '20230724', '20230725', '20230726', '20230727', '20230728', '20230729', '20230730', '20230731', '20230801', '20230802', '20230803', '20230804', '20230805', '20230806', '20230807', '20230808', '20230809', '20230810', '20230811', '20230812', '20230813', '20230814', '20230815', '20230816', '20230817', '20230818', '20230819', '20230820', '20230821', '20230822', '20230823', '20230824', '20230825', '20230826', '20230827', '20230828', '20230829', '20230830', '20230831', '20230901', '20230902', '20230903', '20230904', '20230905', '20230906', '20230907', '20230908', '20230909', '20230910', '20230911', '20230912', '20230913', '20230914', '20230915', '20230916', '20230917', '20230918', '20230919', '20230920', '20230921', '20230922', '20230923', '20230924', '20230925', '20230926', '20230927', '20230928', '20230929', '20230930', '20231001', '20231002', '20231003', '20231004', '20231005', '20231006', '20231007', '20231008', '20231009', '20231010', '20231011', '20231012', '20231013', '20231014', '20231015', '20231016', '20231017', '20231018', '20231019', '20231020', '20231021', '20231022', '20231023', '20231024', '20231025', '20231026', '20231027', '20231028', '20231029', '20231030', '20231031', '20231101', '20231102', '20231103', '20231104', '20231105', '20231106', '20231107', '20231108', '20231109', '20231110', '20231111', '20231112', '20231113', '20231114', '20231115', '20231116', '20231117', '20231118', '20231119', '20231120', '20231121', '20231122', '20231123', '20231124', '20231125', '20231126', '20231127', '20231128', '20231129', '20231130', '20231201', '20231202', '20231203', '20231204', '20231205', '20231206', '20231207', '20231208', '20231209', '20231210', '20231211', '20231212', '20231213', '20231214', '20231215', '20231216', '20231217', '20231218', '20231219', '20231220', '20231221', '20231222', '20231223', '20231224', '20231225', '20231226', '20231227', '20231228', '20231229', '20231230', '20231231', '20240101', '20240102', '20240103', '20240104', '20240105', '20240106', '20240107', '20240108', '20240109', '20240110', '20240111', '20240112', '20240113', '20240114', '20240115', '20240116', '20240117', '20240118', '20240119', '20240120', '20240121', '20240122', '20240123', '20240124', '20240125', '20240126', '20240127', '20240128', '20240129', '20240130', '20240131', '20240201', '20240202'], verbose: False, log: None
Dowloading of the Sentinel-1-Burst-ID map
EGMS-toolkit requires the Sentinel-1-Burst-ID map to detect the datasets. EGMS-toolkit can check the latest version and download the file by using the following method.
By default, the file will be stored in the 3rdparty directory. However, the user can change the directory by modifying the dirmap attributes of the class.
[4]:
info.downloadfile(verbose=False)
[4]:
<EGMStoolkit.classes.EGMSS1burstIDapi.S1burstIDmap at 0x169fde0e0>
Check the tile/bursts available according the user inputs
From the Sentinel-1-Burst-ID map, EGMS-toolkit is now able to detect the associated EGMS datasets. Of course, the user can give some parameters to control the search.
Create the Python variable.
We call the class.
[5]:
ROIpara = EGMSS1ROIapi.S1ROIparameter(verbose=False)
Definition of some parameters
For the EGMS level
The user can define the level of EGMS datasets: i.e., L2a, L2b or L3.
[6]:
ROIpara.egmslevel = 'L2b' # Level of EGMS data
For the ROI (or bbox)
The user must define the ROI. There are three ways to define this parameters:
by a list of EPSG:4326 coordinates [W,S,E,N];
by a country name (i.e., GB, FR, etc.);
by a shapefile in EPSG:4326 coordinates (multiline string format).
[7]:
ROIpara.bbox = [-6.427059639290446,53.2606655698541,-6.0952332730202095,53.41811986118854]
For the release
Currently, there are two available releases: - 2015-2021; - 2018-2022.
The user can select one of these releases.
[8]:
ROIpara.release = '2018_2022'
Conversion of the bbox to a ROI
EGMS-toolkit requires a conversion of the bbox to a ROI. The toolkit will create a shapefile to store the ROI. The default value is bbox.shp.
[9]:
ROIpara.createROI(verbose=False)
[9]:
<EGMStoolkit.classes.EGMSS1ROIapi.S1ROIparameter at 0x169fde020>
Detection of the EGMS datasets
Now, EGMS-toolkit has the required information to detect the EGMS datasets.
By default, all relative orbits and passes will be selected. Of course, the user can used two options to define the relative orbits and pass of Sentinel-1 satellites.
NOTE: Lists of tracks and passes can be given for multiples searches.
[10]:
ROIpara.detectfromIDmap(info,Track_user=1,Pass_user='Ascending',verbose=True)
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.classes.EGMSS1ROIapi.EGMStoolkit.classes.EGMSS1ROIapi:
Detect the data regarding the burst IDs
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSS1ROIapi.py
Copyright 2024, UCD / iCRAG
################################################################################
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSS1ROIapi.checkfile
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSS1ROIapi.py
--> The use of the S1 burst ID map is less accurate than the use of .xml S1 files.
For the L2a and L2b levels
|████████████████████████████████████████| 1127661/1127661 [100%] in 45.1s (24992.20/s)
[10]:
<EGMStoolkit.classes.EGMSS1ROIapi.S1ROIparameter at 0x169fde020>
Save and load results from detection
EGMS-toolkit can save and load the results of a detection.
[11]:
# Save the burst ID list
ROIpara.saveIDlistL2()
# Load the burst ID list
ROIpara.loadIDlistL2()
[11]:
<EGMStoolkit.classes.EGMSS1ROIapi.S1ROIparameter at 0x169fde020>
By default, the file name is egmslist.pkl but the user can change it with the input option.
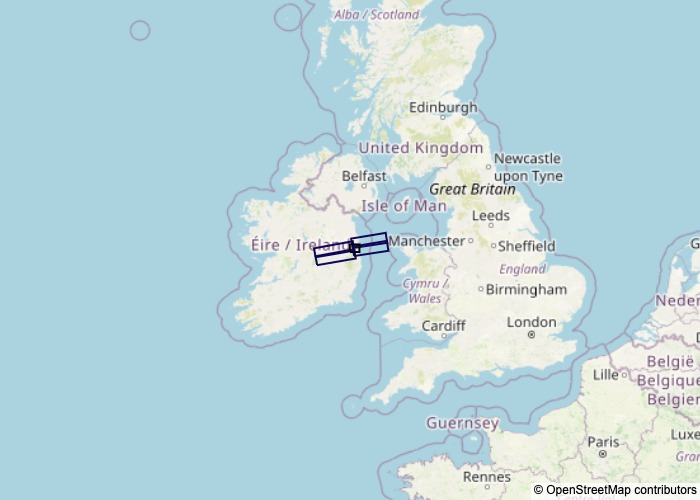
Display a map
To visualise the results, EGMS-toolkit can display the expected datasets.
[12]:
ROIpara.displaymap(output='fig_search.jpg',use_folium=False)
If the ouput option is not given, the figure will be displayed and not saved.
Download the EGMS data
The next step therefore is the downloading of the datasets. However, the user needs to create the links.
Create the python variable
We call the class.
[13]:
downloadpara = EGMSdownloaderapi.egmsdownloader(verbose=False)
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
The WARNING is due to a incorrect token. The user will modify it later.
Update the list of datasets (and create the links)
Now, the user can concatenate their search to create a list of downloading links.
[14]:
downloadpara.updatelist(infoS1ROIparameter=ROIpara)
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
[14]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
And the user can displayed the lists of datasets.
[15]:
downloadpara.printlist(verbose=True)
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.classes.EGMSdownloaderapi.EGMStoolkit.classes.EGMSdownloaderapi:
Print the list(s) of EGMS files
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
Copyright 2024, UCD / iCRAG
################################################################################
For the EGMS data: L2b
File 1: EGMS_L2b_001_0314_IW2_VV_2018_2022_1.zip (Release 2018_2022)
File 2: EGMS_L2b_001_0314_IW3_VV_2018_2022_1.zip (Release 2018_2022)
File 3: EGMS_L2b_001_0315_IW2_VV_2018_2022_1.zip (Release 2018_2022)
File 4: EGMS_L2b_001_0315_IW3_VV_2018_2022_1.zip (Release 2018_2022)
[15]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
Concatenate other searches
Often, the user would like to download datasets with different parameters (i.e., tracks). EGMS-toolkit can concatenate other searches to optimise the downloading time.
Firstly, the next lines will add the search for L3 vertical EGMS datasets of the 2015-2021 release.
[16]:
ROIpara.egmslevel = 'L3'
ROIpara.egmsL3component = 'UD'
ROIpara.release = '2015_2021'
ROIpara.detectfromIDmap(infoburstID=info,verbose=False)
downloadpara.updatelist(infoS1ROIparameter=ROIpara,verbose=False)
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSS1ROIapi.checkfile
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSS1ROIapi.py
--> The use of the S1 burst ID map is less accurate than the use of .xml S1 files.
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
[16]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
Secondly, we will also add an other search for L3 horizontal EGMS datasets of the 2018-2022 release.
[17]:
ROIpara.egmslevel = 'L3'
ROIpara.egmsL3component = 'EW'
ROIpara.release = '2018_2022'
downloadpara.updatelist(infoS1ROIparameter=ROIpara.detectfromIDmap(infoburstID=info))
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSS1ROIapi.checkfile
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSS1ROIapi.py
--> The use of the S1 burst ID map is less accurate than the use of .xml S1 files.
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
[17]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
And the user can print the new list.
[18]:
downloadpara.printlist(verbose=True)
WARNING in EGMS-toolkit processing: in EGMStoolkit.classes.EGMSdownloaderapi.checkparameter
/Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
--> The user token is not correct.
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.classes.EGMSdownloaderapi.EGMStoolkit.classes.EGMSdownloaderapi:
Print the list(s) of EGMS files
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
Copyright 2024, UCD / iCRAG
################################################################################
For the EGMS data: L2b
File 1: EGMS_L2b_001_0314_IW2_VV_2018_2022_1.zip (Release 2018_2022)
File 2: EGMS_L2b_001_0314_IW3_VV_2018_2022_1.zip (Release 2018_2022)
File 3: EGMS_L2b_001_0315_IW2_VV_2018_2022_1.zip (Release 2018_2022)
File 4: EGMS_L2b_001_0315_IW3_VV_2018_2022_1.zip (Release 2018_2022)
For the EGMS data: L3UD
File 1: EGMS_L3_E32N34_100km_U.zip (Release 2015_2021)
For the EGMS data: L3EW
File 1: EGMS_L3_E32N34_100km_E_2018_2022_1.zip (Release 2018_2022)
[18]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
Change the user token
In order to dowload the EGMS data, the user requires to be authorised on the EGMS servers. The hack is to use the user token which can be found, online, at the end of downloading links.
This token is time-limited.
[19]:
downloadpara.token = 'XXXXXXXXXXX'
Download the EGMS data
Now, it is possible to download the data. All the used options are optionnal and are defined by default.
[20]:
downloadpara.download(outputdir='./Output',unzipmode=False,cleanmode=False)
[20]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
Unzip the files
The downlaoded files are .zip archives. The following method will unzip the archives.
However, it is possible to unzip the file directly with the download method by using the unzipmode option.
nbworker option enables the parallel unzipping (number of workers).
[ ]:
downloadpara.unzipfile(outputdir='./Output',unzipmode=True,nbworker=2,cleanmode=True,verbose=True)
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.classes.EGMSdownloaderapi.EGMStoolkit.classes.EGMSdownloaderapi:
Unzip the EGMS files
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/classes/EGMSdownloaderapi.py
Copyright 2024, UCD / iCRAG
################################################################################
1 / 6 files: Unzip the file: EGMS_L3_E32N34_100km_E_2018_2022_1.zip
2 / 6 files: Unzip the file: EGMS_L3_E32N34_100km_U.zip
3 / 6 files: Unzip the file: EGMS_L2b_001_0314_IW3_VV_2018_2022_1.zip
4 / 6 files: Unzip the file: EGMS_L2b_001_0315_IW3_VV_2018_2022_1.zip
5 / 6 files: Unzip the file: EGMS_L2b_001_0315_IW2_VV_2018_2022_1.zip
6 / 6 files: Unzip the file: EGMS_L2b_001_0314_IW2_VV_2018_2022_1.zip
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
Clean the unused files
Then, the user can delete the files that are not in the lists.
[22]:
downloadpara.clean(outputdir='./Output',verbose=False)
[22]:
<EGMStoolkit.classes.EGMSdownloaderapi.egmsdownloader at 0x16a5fe470>
Post-processing of the EGMS data
The files are in the output directory:
├── Output │ ├── L2a │ ├── L2b │ │ ├── 2018_2022 │ │ │ ├── EGMS_L2b_001_0314_IW2_VV_2018_2022_1 │ │ │ │ ├── EGMS_L2b_001_0314_IW2_VV_2018_2022_1.csv │ │ │ │ ├── EGMS_L2b_001_0314_IW2_VV_2018_2022_1.xml │ │ │ ├── EGMS_L2b_001_0315_IW2_VV_2018_2022_1 │ │ │ │ ├── EGMS_L2b_001_0315_IW2_VV_2018_2022_1.csv │ │ │ │ ├── EGMS_L2b_001_0315_IW2_VV_2018_2022_1.xml │ │ │ ├── EGMS_L2b_001_0314_IW3_VV_2018_2022_1 │ │ │ │ ├── EGMS_L2b_001_0314_IW3_VV_2018_2022_1.csv │ │ │ │ ├── EGMS_L2b_001_0314_IW3_VV_2018_2022_1.xml │ │ │ ├── EGMS_L2b_001_0315_IW3_VV_2018_2022_1 │ │ │ │ ├── EGMS_L2b_001_0315_IW3_VV_2018_2022_1.csv │ │ │ │ ├── EGMS_L2b_001_0316_IW3_VV_2018_2022_1.xml │ ├── L3EW │ │ ├── 2018_2022 │ │ │ ├── EGMS_L3_E32N34_100km_E_2018_2022_1 │ │ │ │ ├── EGMS_L3_E32N34_100km_E_2018_2022_1.csv │ │ │ │ ├── EGMS_L3_E32N34_100km_E_2018_2022_1.tiff │ │ │ │ ├── EGMS_L3_E32N34_100km_E_2018_2022_1.xml │ ├── L3UD │ │ ├── 2015_2021 │ │ │ ├── EGMS_L3_E32N34_100km_U │ │ │ │ ├── EGMS_L3_E32N34_100km_U.csv │ │ │ │ ├── EGMS_L3_E32N34_100km_U.tiff │ │ │ │ ├── EGMS_L3_E32N34_100km_U.xml
It therefore is smarter to post-process the files.
Of course, all steps are optional.
Merge the .csv files
The first post-processing is the merging of .csv. The user can define several options: - outputdir the ouput directory; - inputdir the input directory (where are stored the EGMS .csv files); - verbose the verbose mode - paratosave parameter regarding the EGMS names [‘all’ or string value]. [‘latitude’, ‘longitude’, ‘easting’, ‘northing’, ‘height’, ‘height_wgs84’] which will always be saved.
The last option is mode. Indeed, the scripts can find the .csv files based on the search lists (onlist mode) or on the stored files (onfiles mode). For the last mode, the downloadpara class is required.
Three other options are available:
removeduplicate (bool): Reduce the duplicate points [Default: True]
length_threshold (int): Length for the concave hull [Default: 1000]
usevrtmerging (bool): Use the vrt for merging [Default: False]
[23]:
egmsdatatools.datamergingcsv(infoEGMSdownloader=downloadpara,inputdir='./Output',outputdir='./Output',mode='onlist',verbose=True,paratosave='all')
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.functions.egmsdatatools.EGMStoolkit.functions.egmsdatatools:
Merge the datasets in csv format
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/functions/egmsdatatools.py
Copyright 2024, UCD / iCRAG
################################################################################
Output Directory: ./Output
Input Directory: ./Output
Selected parameters: all
Mode: onlist
Merging for EGMS_L3_2015_2021_UD...
Merging for EGMS_L2b_001_VV_2018_2022_1...
Merging for EGMS_L3_2018_2022_1_EW...
Merge the .tiff files (only for the vertical and horizontal displacements)
The .tiff can also be merged. The options are similar.
The paratosave option does not exist here.
[26]:
egmsdatatools.datamergingtiff(infoEGMSdownloader=downloadpara,inputdir='./Output',outputdir='./Output',mode='onlist',verbose=True)
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.functions.egmsdatatools.EGMStoolkit.functions.egmsdatatools:
Merge the datasets in tiff format (only for L3 level)
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/functions/egmsdatatools.py
Copyright 2024, UCD / iCRAG
################################################################################
Output Directory: ./Output
Input Directory: ./Output
Selected parameters: all
Mode: onlist
Merging for EGMS_L3_2015_2021_UD...
Used command: gdal_merge.py -o ./Output/EGMS_L3_2015_2021_UD.tiff -n -9999 -a_nodata -9999 ./Output/L3UD/2015_2021/EGMS_L3_E32N34_100km_U/EGMS_L3_E32N34_100km_U.tiff
0...10...20...30...40...50...60...70...80...90...100 - done.
Merging for EGMS_L3_2018_2022_1_EW...
Used command: gdal_merge.py -o ./Output/EGMS_L3_2018_2022_1_EW.tiff -n -9999 -a_nodata -9999 ./Output/L3EW/2018_2022/EGMS_L3_E32N34_100km_E_2018_2022_1/EGMS_L3_E32N34_100km_E_2018_2022_1.tiff
0...10...20...30...40...50...60...70...80...90...100 - done.
Clip/crop the data
The user can clip or crop the files regarding the ROI (or another shapefile).
The new parameter namefile allows to define the file. By default, ‘all’ will clip/crop all the files. The option clipuseogr2ogr (bool, optional) can be used to force the use of ogr2ogr for clipping (default is False).
[27]:
egmsdatatools.dataclipping(inputdir='./Output',outputdir='./Output',namefile='all',shapefile='bbox.shp',verbose=True)
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.functions.egmsdatatools.EGMStoolkit.functions.egmsdatatools:
Clip the dataset(s)
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/functions/egmsdatatools.py
Copyright 2024, UCD / iCRAG
################################################################################
The file name is: all
Input Directory: ./Output
Output Directory: ./Output
Shapefile: bbox.shp
1 / 5 file(s): Clip the file ./Output/EGMS_L3_2015_2021_UD.csv to ./Output/EGMS_L3_2015_2021_UD_clipped.csv...
2 / 5 file(s): Clip the file ./Output/EGMS_L3_2018_2022_1_EW.csv to ./Output/EGMS_L3_2018_2022_1_EW_clipped.csv...
3 / 5 file(s): Clip the file ./Output/EGMS_L2b_001_VV_2018_2022_1.csv to ./Output/EGMS_L2b_001_VV_2018_2022_1_clipped.csv...
4 / 5 file(s): Crop the file ./Output/EGMS_L3_2015_2021_UD.tiff to ./Output/EGMS_L3_2015_2021_UD_cropped.tiff...
5 / 5 file(s): Crop the file ./Output/EGMS_L3_2018_2022_1_EW.tiff to ./Output/EGMS_L3_2018_2022_1_EW_cropped.tiff...
Clean the raw files and directories
To save space, the raw files and directories can be deleted.
[28]:
egmsdatatools.removerawdata(inputdir='./Output',verbose=True)
################################################################################
EGMStoolkit 0.2.1 Beta
--------------------------------------------------------------------------------
EGMStoolkit.functions.egmsdatatools.EGMStoolkit.functions.egmsdatatools:
Delete the raw dataset(s) from EGMS
Script: /Users/alexis_hrysiewicz/Work/UCD_InSAR_Scripts/EGMStoolkit/src/EGMStoolkit/functions/egmsdatatools.py
Copyright 2024, UCD / iCRAG
################################################################################
Input Directory: ./Output
Deleting...
Deleting...
Post-processing for advanced users
The next sections show some other commands to post-process the data.
Interpolation of point data (.csv) into a .tif raster file
The first command is the possibility to interpolate the point data (from .csv files) into a .tif raster file. Of course, this step can be can be done before the cropping/clipping step.
Creation of the dict. for the gridding parameters
The user requires to define a dictionary with the gridding parameter.
[32]:
paragrid = dict()
paragrid['Xmin'] = 2896000 # Minimal X coordinate in EPGS:3035
paragrid['Ymin'] = 3317250 # Minimal Y coordinate in EPGS:3035
paragrid['Xmax'] = 3359000 # Maximal X coordinate in EPGS:3035
paragrid['Ymax'] = 3745500 # Maximal Y coordinate in EPGS:3035
paragrid['xres'] = 50 # X spatial resolution in EPGS:3035
paragrid['yres'] = 50 # Y spatial resolution in EPGS:3035
paragrid['algo'] = 'average:radius1=50:radius2=50:angle=0.0:nodata=-9999' # Alfgorithm used and options
paragrid['variable'] = 'mean_velocity,mean_velocity_std,acceleration,acceleration_std,seasonality,seasonality_std'
# paragrid['algo'] = 'invdist:power=2.0:smoothing=0.0:radius1=0.0:radius2=0.0:angle=0.0:max_points=0:min_points=0:nodata=0.0'
# paragrid['algo'] = 'invdistnn:power=2.0:radius=1.0:max_points=12:min_points=0:nodata=0'
# paragrid['algo'] = 'average:radius1=0.0:radius2=0.0:angle=0.0:min_points=0:nodata=0.0'
# paragrid['algo'] = 'nearest:radius1=0.0:radius2=0.0:angle=0.0:nodata=0.0'
# paragrid['algo'] = 'linear:radius=-1.0:nodata=0.0'
And the user can run the interpolation.
[ ]:
egmsdatatools.datagridding(inputdir='./Output',outputdir='./Output',namefile='all',verbose=True,paragrid=paragrid)
Conversion of .csv files to other vector format
The last command will convert the .csv files to other vector formats. The available formats are:
ESRI Shapefile (default);
GPKG;
GeoJSON.
[ ]:
egmsdatatools.convertcsv(verbose=False)